

Ph.D. Candidate
 National University of Singapore
National University of SingaporeI am a Ph.D. student at the National University of Singapore working in computational and cognitive neuroscience. My research focuses on the neural mechanisms 🧠 underlying learning, memory, and decision-making, and I am interested in how physiological factors influence the acquisition and exploitation of knowledge. I develop interpretable computational models 🖥️ grounded in theory and experimental data to explain and predict cognitive processes and behavior.
Currently, I am building biologically plausible models of spatial learning and associative memory, advised by Andrew Tan, Camilo Libedinsky, and Shih-Cheng Yen. Previously, I worked on reinforcement learning and fMRI research Gedi Lukšys, and contributed to monkey connectome research with Anna Wang Roe.
Warning
Problem: The current name of your GitHub Pages repository ("Solution: Please consider renaming the repository to "
http://".
However, if the current repository name is intended, you can ignore this message by removing "{% include widgets/debug_repo_name.html %}" in index.html.
Action required
Problem: The current root path of this site is "baseurl ("_config.yml.
Solution: Please set the
baseurl in _config.yml to "Education
-
 National University of SingaporePh.D. Student in MedicineNeuroscience and Biostatistics dual Track2023 - present
National University of SingaporePh.D. Student in MedicineNeuroscience and Biostatistics dual Track2023 - present -
 Zhejiang UniversityB.Sc. in Bioinformatics2019 - 2023
Zhejiang UniversityB.Sc. in Bioinformatics2019 - 2023 -
 The University of EdinburghB.Sc. (Hons) in Biomedical Informatics2019 - 2023
The University of EdinburghB.Sc. (Hons) in Biomedical Informatics2019 - 2023
Experience
-
 National University of SingaporeCamilo Libedinsky lab Graduate ResearcherSep. 2023 - present
National University of SingaporeCamilo Libedinsky lab Graduate ResearcherSep. 2023 - present -
 The University of EdinburghGediminas Lukšys lab Research InternMar. 2022 - Aug. 2023
The University of EdinburghGediminas Lukšys lab Research InternMar. 2022 - Aug. 2023 -
 Zhejiang UniversityZhiping Wang lab Research InternJan. 2022 - Sep. 2022Jun. 2020 - Aug. 2020
Zhejiang UniversityZhiping Wang lab Research InternJan. 2022 - Sep. 2022Jun. 2020 - Aug. 2020 -
 Zhejiang UniversityAnna Wang Roe lab Research InternApr. 2021 - Oct. 2021
Zhejiang UniversityAnna Wang Roe lab Research InternApr. 2021 - Oct. 2021
Honors & Awards
-
NUS Research Scholarship
-
Outstanding Graduate of Zhejiang University
-
Zhejiang University Scholarship
-
ZJU-UoE Institute Academic Scholarship (¥40,000)
News
Selected Publications (view all )

A biologically plausible computational model of hippocampal neurogenesis and pattern separation in memory
Jiachuan Wang, Vachan Shetru Jagadeesh, M Ganesh Kumar, Camilo Libedinsky, Shih-Cheng Yen, Andrew Yong-Yi Tan, Jai S Polepalli
Cognitive Computational Neuroscience (CCN) 2025
We developed a computational model incorporating an entorhinal cortex-dentate gyrus circuit. Simulations suggested that impaired synaptic plasticity or increased neurogenesis alone was sufficient to disrupt behavioral pattern separation.
A biologically plausible computational model of hippocampal neurogenesis and pattern separation in memory
Jiachuan Wang, Vachan Shetru Jagadeesh, M Ganesh Kumar, Camilo Libedinsky, Shih-Cheng Yen, Andrew Yong-Yi Tan, Jai S Polepalli
Cognitive Computational Neuroscience (CCN) 2025
We developed a computational model incorporating an entorhinal cortex-dentate gyrus circuit. Simulations suggested that impaired synaptic plasticity or increased neurogenesis alone was sufficient to disrupt behavioral pattern separation.
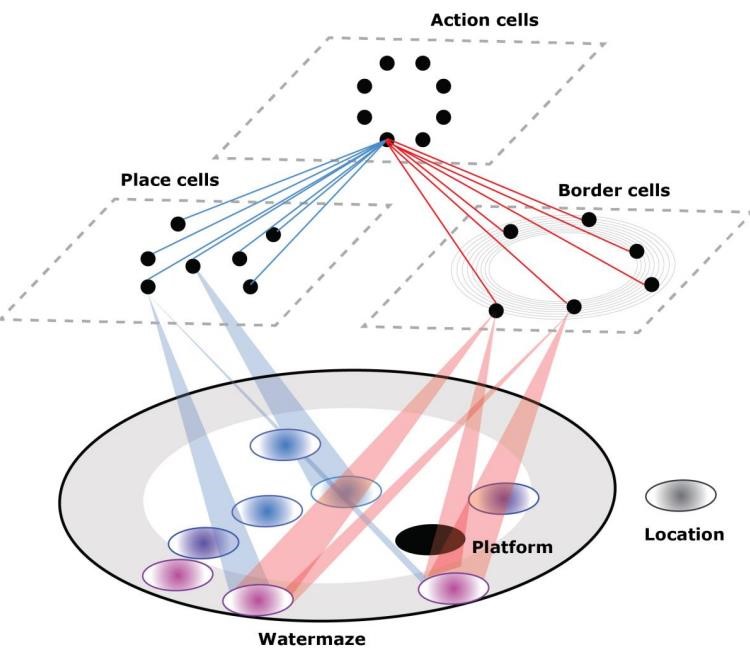
Computational model-based analysis of spatial navigation strategies under stress and uncertainty using place, distance and border cells
Shiqi Wang, Jiachuan Wang, Wenyuan Zhu, Yuchen Cheng, Beste Aydemir, Yanran Qiu, Wulfram Gerstner, Carmen Sandi, Gedi Luksys
Neuroscience 2023: Society for Neuroscience (SfN) 2023
We investigated how stress and uncertainty affect learning in two mouse strains using the Morris water maze and modeled their behavior with a biologically plausible network of border and place cells, revealing computational correlates of temperature-based differences.
Computational model-based analysis of spatial navigation strategies under stress and uncertainty using place, distance and border cells
Shiqi Wang, Jiachuan Wang, Wenyuan Zhu, Yuchen Cheng, Beste Aydemir, Yanran Qiu, Wulfram Gerstner, Carmen Sandi, Gedi Luksys
Neuroscience 2023: Society for Neuroscience (SfN) 2023
We investigated how stress and uncertainty affect learning in two mouse strains using the Morris water maze and modeled their behavior with a biologically plausible network of border and place cells, revealing computational correlates of temperature-based differences.
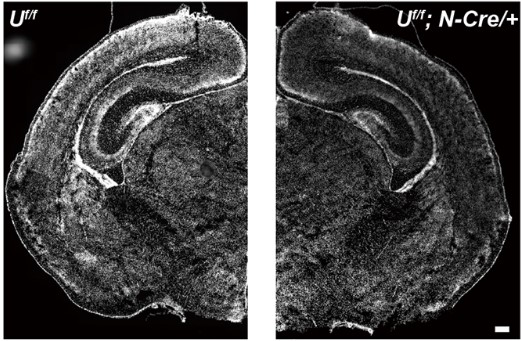
Fine-tuning of mTOR signaling by the UBE4B-KLHL22 E3 ubiquitin ligase cascade in brain development
Xiangxing Kong, Xin Shu, Jiachuan Wang, Dandan Liu, Yingchun Ni, Weiqi Zhao, Lebo Wang, Zhihua Gao, Jiadong Chen, Bing Yang, Xing Guo, Zhiping Wang
Development 2022
The UBE4B-KLHL22 E3 ubiquitin ligase cascade regulates mTOR activity in neurodevelopment, and its disruption impairs neural precursor proliferation and differentiation, which can be rescued by mTOR inhibition.
Fine-tuning of mTOR signaling by the UBE4B-KLHL22 E3 ubiquitin ligase cascade in brain development
Xiangxing Kong, Xin Shu, Jiachuan Wang, Dandan Liu, Yingchun Ni, Weiqi Zhao, Lebo Wang, Zhihua Gao, Jiadong Chen, Bing Yang, Xing Guo, Zhiping Wang
Development 2022
The UBE4B-KLHL22 E3 ubiquitin ligase cascade regulates mTOR activity in neurodevelopment, and its disruption impairs neural precursor proliferation and differentiation, which can be rescued by mTOR inhibition.