LTR predictor: HERV-oriented alignment tool
A BLAST-like algorithm, which can perform fast alignment of repeats while supporting custom input of reference sequences.
This was an undergraduate course assignment and now I am no longer working on genetics.
The project was selected as the top-5 mini-projects of Biomedical Informatics 3 (BMI3) coursework. https://labw.org/bmi3-2021/miniprojects.
Introduction
The human genome is rich in retroviruses and retroviral elements integrated during the evolutionary process1. Among human endogenous retroviruses (HERVs), HERV-K is the most transcriptionally active group1. It plays a vital role in embryogenesis, whereas closely related to cancer and neurodegenerative diseases234. (Grow et al., 2015, Li et al., 2015, Argaw-Denboba et al., 2017). HERVs are repeats with low complexity, making it hard to perform annotation and analysis geared to the genome5. Algorithms like cross_match and WindowMasker used to find repeats masked a large quantity of the annotated exons 5. Here, we have developed a basic local alignment search tool (BLAST)-like algorithm named the LTR predictor, which can perform fast alignment of repeats while supporting custom input of reference sequences. Our test on LTR5_Hs (a type of HERV-K) shows that the LTR predictor has good accuracy and running speed, and can provide inspiration for predicting the genome coordinates of repeat such as HERV. The HERV consensus sequences (DF0000471, DF0000472, and DF0000558) were downloaded from Dfam database (https://dfam.org/home). Soft-masked reference sequences of human genome (GRCh38 Genome Reference Consortium Human Reference 38, or hg38) were from UCSC Genome browser (https://genome.ucsc.edu/).
Algorithm Implementation
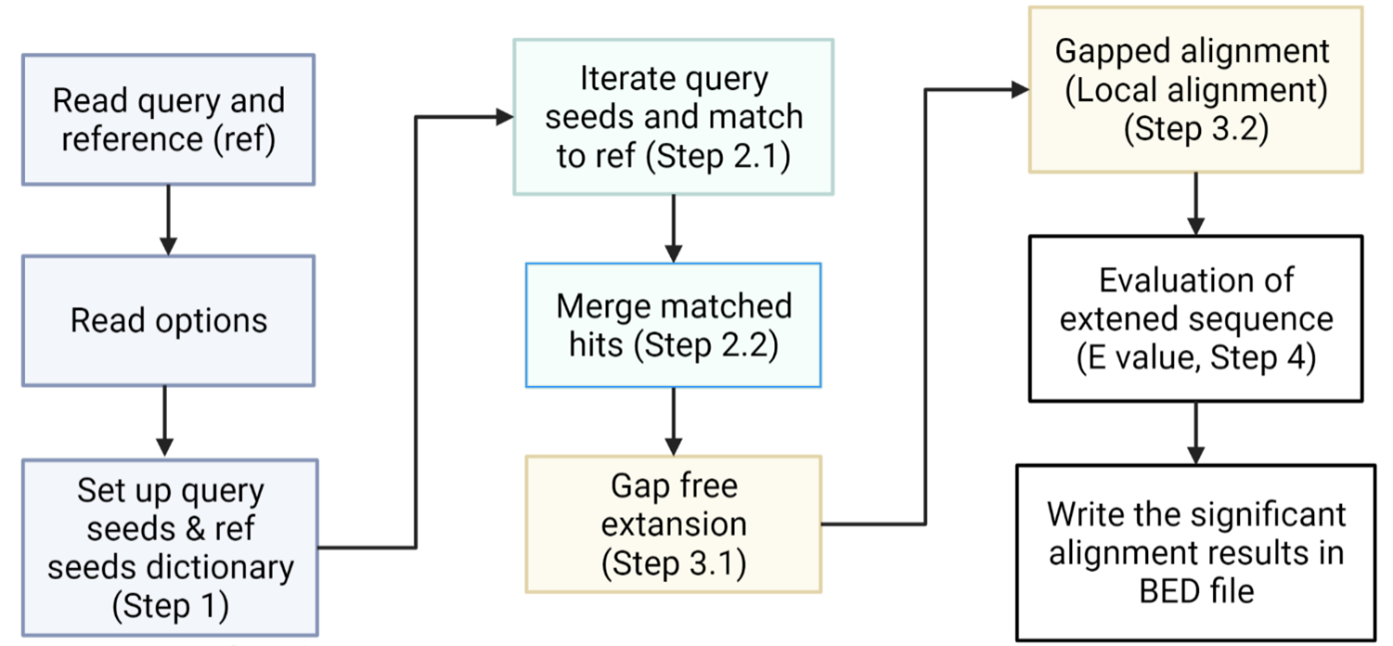
Aiming at identifying the long terminal repeated (LTR) coordinates on the reference chromosome, our algorithm allows users to one query file (query) and one reference chromosome file (ref) in FASTA format. It returns a BED file containing the start and end indexes on the reference chromosome with its name, on which the query sequence aligned (Fig. 1). The query file could be any LTR sequence from Dfam or other databases and it is recommended that the ref is in soft-masked or unmasked format, which provides the sequence information of LTR rather than masks them. The core function is based on the Smith-Waterman algorithm to achieve local alignment with dynamic programming.
Our LTR finder has four major steps including seeding, hits calling, extending, and evaluation. After evaluation, statistically significant alignment results are written into a BED file with the chromosome name, start and end indexes of alignment. The overall workflow is listed step by step with a general flow chart (Fig. 1).
Overall Workflow
- Read in the query and ref in FASTA format and record the optional parameters;
- Generate query and ref seeds of length 11-nucleotide and save them in a dictionary, where keys are the seed sequence and values are indexes on query and ref, respectively;
- Read in the query and ref in FASTA format and record the optional parameters;
- Generate query and ref seeds of length 11-nucleotide and save them in a dictionary, where keys are the seed sequence and values are indexes on query and ref, respectively;
- Find exactly matched seeds as hits using the property of the key in the dictionary;
- Merge overlapped and nearby hits within mismatch threshold (-m);
- Run a gap free extension on both sides of hits using hamming distance like score methods;
- Local alignment of sequences after gap free extension by Smith-Waterman algorithm and deliver an alignment score to each matched sequence;
- Evaluate the E-score after local alignment to reduce the false positive rate;
- Write the chromosome name, start and end coordinates into a BED file.
Source Code
https://github.com/Haoninghui/BMI3_Project1
Acknowledgements
This project was a group work of Biomedical Informatics 3 course with Hao Ninghui. Architecture and acceleration were aided and advised by Yu Zhejian. Many thanks. The program tests used the analysis server of ZJE Institute. Thanks for the guidance and feedback from Dr. Wanlu Liu, Dr. Hugo Samano-Sanchez, and teaching assistant Ziwei Xue.
References
GARCIA-MONTOJO, M., DOUCET-O’HARE, T., HENDERSON, L. AND NATH, A. (2018) Human endogenous retrovirus-K (HML-2): a comprehensive review., Critical reviews in microbiology, 44(6), pp. 715-738. doi: 10.1080/1040841X.2018.1501345. ↩︎ ↩︎2
GROW, E. J., FLYNN, R. A., CHAVEZ, S. L., BAYLESS, N. L., WOSSIDLO, M., WESCHE, D. J., MARTIN, L., WARE, C. B., BLISH, C. A., CHANG, H. Y., PERA, R. A. R. AND WYSOCKA, J. (2015) Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells., Nature, 522(7555), pp. 221-225. doi: 10.1038/nature14308. ↩︎
LI, W., LEE, M.-H., HENDERSON, L., TYAGI, R., BACHANI, M., STEINER, J., CAMPANAC, E., HOFFMAN, D. A., VON GELDERN, G., JOHNSON, K., MARIC, D., MORRIS, H. D., LENTZ, M., PAK, K., MAMMEN, A., OSTROW, L., ROTHSTEIN, J. AND NATH, A. (2015) Human endogenous retrovirus-K contributes to motor neuron disease., Science translational medicine, 7(307), p. 307ra153. doi: 10.1126/scitranslmed.aac8201. ↩︎
ARGAW-DENBOBA, A., BALESTRIERI, E., SERAFINO, A., CIPRIANI, C., BUCCI, I., SORRENTINO, R., SCIAMANNA, I., GAMBACURTA, A., SINIBALDI-VALLEBONA, P. AND MATTEUCCI, C. (2017) HERV-K activation is strictly required to sustain CD133+ melanoma cells with stemness features., Journal of experimental & clinical cancer research: CR, 36(1), p. 20. doi: 10.1186/s13046-016-0485-x. ↩︎
LI, X., KAHVECI, T. AND SETTLES, A. M. (2008) A novel genome-scale repeat finder geared towards transposons., Bioinformatics (Oxford, England). England, 24(4), pp. 468-476. doi: 10.1093/bioinformatics/btm613. ↩︎ ↩︎2